Abstract
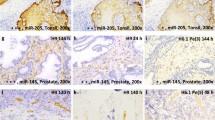
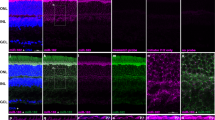
There are relatively few protocols described for the in situ detection of microRNA (miRNA) and they often use cryostat sections, signal amplification and hybridization or washes of 50–60 °C. This protocol describes in situ miRNA detection that can be done in paraffin-embedded, formalin-fixed tissue. Detection of the miRNA precursors can be done by RT in situ PCR, which can theoretically detect one copy per cell. The key variable for the RT in situ PCR protocol is optimal protease digestion, which is then followed by overnight DNase digestion and target specific incorporation of the reported nucleotide into the amplified cDNA. Detection of mature miRNAs is achieved by in situ hybridization with locked nucleic acid probes. This part of the protocol involves a brief protease digestion, followed by an overnight hybridization, short low stringency wash and detection of the labeled probe. The key variables for this method include probe concentration and stringency conditions. Each miRNA in situ method takes 1 d. The final step of the protocol involves colabeling by immunohistochemistry for the putative target of the miRNA, which is done after the in situ hybridization step and takes a few hours.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
25 March 2010
In the version of this article initially published, the following text was omitted from the acknowledgments section: “We also thank Nicola Valeri, Pierliugi Gasparini and Muller Fabbri for their excellent work in providing the basis for the bcl-2/miR-16, MSH 2/miR-155 and miR-155 and Castleman’s disease associations that were an important part of the colabeling experiments. Dr Valeri is a Fellow of the American-Italian Cancer Foundation.” In addition, in Table 1, the information in the fifth row (for ref. 11) is incorrect, as described below: • In the “Probe label” column, “FITC” should be “FITC or Cy3”. • In the “Probe concentration” column, “Not listed” should be “2.5 pmol per 20 μl”. • In the “Wash” column, “37 °C, not listed” should be “37 °C, 1× SSC”. • In the “Tissue” column, “FFPE” should be “PFFCC”. • In the “Colabel/RT in situ PCR” column, “”No/No” should be “Yes/No”. Accordingly, the definition for PFFCC (permeabilized, formalin-fixed cells on coverslips) has been added to the table footnote. These errors have been corrected in the HTML and PDF versions of the article.
References
Nuovo, G.J. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods 44, 39–46 (2008).
Lee, E.J. et al. Classification of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14, 35–42 (2008).
Calin, G.A. & Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 (2006).
Garzon, R., Fabbri, M., Cimmino, A., Calin, G.A. & Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 12, 580–587 (2006).
Yanaihara, N. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 (2006).
Gramantieri, L. et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 67, 6092–6099 (2007).
Cimmino, A. et al. miR-15 and miR-16 induce apoptosis by targeting bcl-2. Proc. Natl. Acad. Sci. USA 102, 13944–13949 (2005).
Kuhn, D.E. et al. Experimental validation of miR targets. Methods 44, 47–54 (2008).
Nuovo, G.J. Co labeling using RT in situ PCR: a review. J. Histochem. Cytochem. 49, 1329–1339 (2001).
Nuovo, G.J. PCR In Situ Hybridization: Protocols and Applications 3rd edn. (Lippincott, Williams and Wilkins, Raven Press, New York, 1996).
Politz, J.C., Zhang, F. & Pederson, T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. USA 103, 18957–18962 (2006).
Obernosterer, G., Martinez, J. & Alenius, M. Locked nucleic acid based in situ detection of microRNAs in mouse tissue sections. Nat. Protoc. 2, 1508–1514 (2007).
Sempere, L.F. et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 67, 11612–11620 (2007).
Schetter, A.J. et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299, 425–436 (2008).
Silahtaroglu, A.N. et al. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat. Protoc. 2, 2520–2528 (2007).
Bak, M. et al. MicroRNA expression in the adult mouse nervous system. RNA 14, 432–444 (2008).
Jiang, J., Lee, E.J., Gusev, Y. & Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 33, 5394–5403 (2005).
Schmittgen, T.D., Jiang, J., Liu, Q. & Yang, L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 32, 43–47 (2004).
Liang, Y., Ridzon, D., Wong, L. & Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166 (2007).
Acknowledgements
We greatly appreciate the assistance and reagents from Ventana Medical Systems, and the direct assistance of Christopher Roberts, PhD, Kathleen Sergott and Margaret Nuovo, MD, who assisted with the photography and the figures. This work was supported by a grant from the Lewis Foundation (G.J.N.) and R21 CA114304 (T.D.S.).
We also thank Nicola Valeri, Pierliugi Gasparini and Muller Fabbri for their excellent work in providing the basis for the bcl-2/miR-16, MSH 2/miR-155 and miR-155 and Castleman’s disease associations that were an important part of the colabeling experiments. Dr Valeri is a Fellow of the American-Italian Cancer Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nuovo, G., Elton, T., Nana-Sinkam, P. et al. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc 4, 107–115 (2009). https://doi.org/10.1038/nprot.2008.215
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.215
This article is cited by
-
A novel approach for microRNA in situ hybridization using locked nucleic acid probes
Scientific Reports (2021)
-
Dicer generates a regulatory microRNA network in smooth muscle cells that limits neointima formation during vascular repair
Cellular and Molecular Life Sciences (2017)
-
Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer
Cell Death & Disease (2016)
-
In Vivo Detection of miRNA Expression in Tumors Using an Activatable Nanosensor
Molecular Imaging and Biology (2016)
-
Micro RNAs: The Future of Idiopathic Pulmonary Fibrosis Therapy
Cell Biochemistry and Biophysics (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.