Abstract
Total serum bilirubin is associated with several clinical outcomes, including cardiovascular disease, diabetes and drug metabolism. We conducted a genome-wide association study in 619 healthy unrelated African Americans in an attempt to replicate reported findings in Europeans and Asians and to identify novel loci influencing total serum bilirubin levels. We analyzed a dense panel of over two million genotyped and imputed SNPs in additive genetic models adjusting for age, sex, and the first two significant principal components from the sample covariance matrix of genotypes. Thirty-nine SNPs spanning a 78 kb region within the UGT1A1 displayed P-values <5 × 10−8. The lowest P-value was 1.7 × 10−22 for SNP rs887829. None of SNPs in the UGT1A1 remained statistically significant in conditional association analyses that adjusted for rs887829. In addition, SNP rs10929302 located in phenobarbital response enhancer module was significantly associated with bilirubin level with a P-value of 1.37 × 10−11; this enhancer module is believed to have a critical role in phenobarbital treatment of hyperbilirubinemia. Interestingly, the lead SNP, rs887829, is in strong linkage disequilibrium (LD) (r2≥0.74) with rs10929302. Taking advantage of the lower LD and shorter haplotypes in African-ancestry populations, we identified rs887829 as a more refined proxy for the causative variant influencing bilirubin levels. Also, we replicated the reported association between variants in SEMA3C and bilirubin levels. In summary, UGT1A1 is a major locus influencing bilirubin levels and the results of this study promise to contribute to understanding of the etiology and treatment of hyperbilirubinaemia in African-ancestry populations.
Similar content being viewed by others
INTRODUCTION
Bilirubin is an end product of heme metabolism. Heme, a component of hemoglobin in red blood cells, is broken down by heme oxygenase resulting in bilirubin.1, 2 Glucuronidation by UDP-glycosyltransferase in hepatocytes3 is an important step in facilitating the excretion of bilirubin into bile.4 A very high level of bilirubin, especially in children, is neurotoxic. In contrast and because of its potent antioxidant effect, moderately elevated bilirubin has been shown to be protective against several adult oxidative stress-mediated diseases including diabetes mellitus, diabetic nephropathy, cancer and cardiovascular disease.5, 6, 7, 8, 9 Moderately higher levels of bilirubin within the normal range have been associated with reduced risk of respiratory disease and all cause mortality.10 Additionally, high serum bilirubin is apparently beneficial for survival in the elderly population.11
Previously, linkage studies identified a serum bilirubin QTL on chromosome 2q (containing the UDP glycosyltransferase 1 family, UGT1A) in European-ancestry populations, a finding that was replicated in American Indians.12, 13, 14 Recently, multiple genome-wide association studies (GWAS) identified variants in UGT1A to be associated with serum bilirubin levels in European- and East-Asian-ancestry populations.15, 16, 17 The aim of the present study is to investigate if these findings extend to African-ancestry populations, and to identify novel loci by conducting a GWAS and replication study of serum bilirubin in apparently healthy unrelated African Americans. Multiple studies have reported that African-ancestry populations have lower bilirubin levels compared with other ancestral groups.18, 19 Understanding the genetic and non-genetic basis of circulating levels of bilirubin is particularly important in populations of African ancestry given the reported higher prevalence of low bilirubin related health conditions (eg, diabetes, the heart and kidney diseases) in African Americans.
Materials and methods
Ethics statement
Ethical approval for the Howard University Family Study (HUFS) was obtained from the Howard University IRB and written informed consent was obtained from each participant.
Study sample
The HUFS is a population-based family study of African Americans in the Washington, D.C. metropolitan area.20 The major objective of the HUFS was to enroll and examine a randomly ascertained sample of African American families (ie, pedigree data collected), along with a set of unrelated individuals (no pedigree data collected), from the Washington, D.C. metropolitan area for the study of the genetic and environmental bases of common complex diseases including hypertension, obesity, diabetes and associated phenotypes. In order to maximize the utility of this sample for the study of multiple common traits, families were not ascertained based on any phenotypes. To estimate heritability of serum bilirubin, we used 1314 individuals within 328 pedigrees from HUFS. Heritability was estimated using SOLAR.21 For the GWAS, we included the subset of apparently healthy individuals (defined as no type 2 diabetes, no hypertension and no other major morbidity) with bilirubin measurements and Affymetrix 6.0 (Santa Clara, CA, USA) SNP genotype data. The 619 individuals that met these criteria comprised 330 subjects enrolled as individuals and 289 unrelated subjects from families (1 randomly selected from related persons in each pedigree).
Serum total bilirubin measurement
Total bilirubin was measured on a Cobas Integra 400 Plus (Roche Diagnostics, Indianapolis, IN, USA) using the Diazo method. Briefly, this method is a colorimetric method based on the reaction between bilirubin and diazotized sulfanilic acid solution to produce a complex known as azobilirubin whose maximum absorbance is pH dependant; to preserve the pH of the reaction an oxalic acid/ sulfanilic acid buffer is used. The color intensity of the reaction is proportional to the concentration of total bilirubin present in the sample and is determined by the increase in absorbance at 552 nm. The minimum detection level for this assay is 0.099 mg/dl. The reproducibility of the assay measured by the coefficient of variation between and within assays is between 0.45 and 0.8%. Serum total bilirubin is reported in mg/dl.
Genotyping
DNA was extracted using buffy coat samples following the manufacturer's instructions using a Gentra PUREGENE DNA Isolation Kit (QIAGEN, Valencia, CA, USA). After sample processing, genome-wide genotyping was performed using the Affymetrix Genome Wide Human SNP Array 6.0.22 Genotypes were called using Birdseed, version 2.22 All samples passed a sample success rate of 95%. SNPs were excluded if they had a success rate of <95% (41 885 SNPs excluded), a minor allele frequency (MAF) ≤0.01 (19 154 SNPs excluded), or had a P-value for the Hardy-Weinberg test of equilibrium <10−3 (6317 SNPs excluded). The current analysis focused on the 808 465 autosomal SNPs that passed these filters. The sample genotyping rate for this set of SNPs in these individuals was 99.5%. The concordance of blind duplicates was 99.74%. In addition, imputation was performed as previously reported.23 Briefly, we successfully imputed 1 506 100 autosomal SNPs using the YRI reference panel and an additional 52 291 SNPs using the CEU reference panel, for a total of 2 366 856 experimentally determined and imputed SNPs.
Check for population stratification
EIGENSTRAT24 was used to detect and correct for population stratification. This method is based on principal components analysis and the resulting correction is specific to a candidate marker's variation in frequency across ancestral populations, minimizing spurious associations whereas maximizing power to detect true associations. The first two principal components were significant and were included as covariates in the regression analysis.
Association and replication analyses
All association and conditional association testing was performed using PLINK.25 Serum bilirubin levels were natural log transformed to follow a normal distribution. Association between an autosomal marker and serum bilirubin was assessed using linear regression assuming an additive genetic effect model with adjustments for age, sex and first two principal components from EIGENSTRAT. We estimated the genomic inflation factor (λGC) based on the median chi-squared test of all study participants.26 Conditional analyses to examine specific SNP associations were performed using PLINK by specifying the associated SNP as a covariate along with the previously mentioned covariates in the linear regression model.
We attempted to replicate published GWAS of serum bilirubin15, 16, 17 in the HUFS samples in two stages: direct (ie, same SNPs as reported) and local (ie, SNPs in reference ancestry linkage disequilibrium (LD) with reported SNPs) replication as previously reported.27 Of the 13 reported SNPs, we were able to directly test 7 in this study; the remaining 6 were either not present on the Affymetrix 6.0 chip, were not successfully imputed or failed our quality control filters. Local replication was performed for SNPs not directly replicated. The SNPs in LD at an r2≥0.3 with discovery variants were queried. HapMap CEU reference data for European-ancestry populations and the HapMap CHB data for the East Asian-ancestry populations were used in the local replication analysis. To adequately account for multiple testing, we estimated the effective degrees of freedom for the spectrally decomposed covariance matrix for the block of SNPs using the HUFS genotyped SNP data as previously described.27, 28
Power calculations were done using the Quanto software package.29 We used a mean (SD) of serum bilirubin=0.58 (0.31) mg/dl, MAF values obtained from HUFS genotype dataset, and assumed an additive genetic model with α=5 × 10−8 in two tail tests to calculate study power.
Results
Clinical characteristics for the 619 participants included in this study are presented in Table 1. Consistent with the national data from NHANES III, male participants had significantly higher levels of bilirubin than the females.30 In contrast, female participants were heavier and had higher levels of insulin, and male participants had higher systolic blood pressure, fasting glucose and alanine amino transferase levels. Only a small proportion (3.8%) of the HUFS participants had elevated (ie, >1.3 mg/dl) serum total bilirubin level. The heritability (h2) of serum total bilirubin in HUFS is 0.42 (SE 0.064) with a P-value of 6.63 × 10−13.
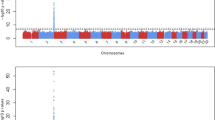
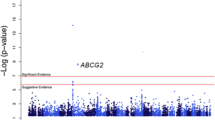
The distribution of genome-wide association P-values for total serum bilirubin is displayed in a Manhattan plot (Supplementary Figure 1) and in a quantile-quantile plot (Supplementary Figure 2). The value of genomic control inflation factor (λGC) was 1.008, which indicated that inflation of association test statistics because of population stratification was negligible. Out of the 100 top scoring SNPs 39 (Supplementary Table S1) were located within a 78 kb region in the UGT1A1 gene on 2q37; all 39 SNPs displayed P-values lower than the predetermined genome wide threshold of 5 × 10−8 (Supplementary Figure 3 and Supplementary Table S2). The lowest P-value corresponded to rs887829 (1.97 × 10−22). After conditioning on the most significant SNP (rs887829), none of the remaining 38 SNP in the UGT1A1 gene maintained genome-wide significance (Supplementary Table S2). This finding is consistent with the observation that the 38 SNPs are in moderate to strong LD with rs887829 (r2 ranged from 0.28 to 1.0 in this study). Of note, previous studies identified rs887829 as a modifier of bilirubin levels with each copy of the T allele resulting in an average increase of 1.77 mg/dl in serum bilirubin in Europeans;17, 31 the corresponding estimate of effect size in this study of African Americans is 1.25 mg/dl per copy of the T allele. SNP rs887829 explained 12.4% of the variance in serum total bilirubin in this study of African Americans.
We directly replicated the top scoring SNP (rs11891311, P-value=4.78 × 10−156) reported in the Korean GWAS.15 This SNP was also reported in the GWAS conducted in European-ancestry populations;16, 17 however, SNP rs11891311 is in moderate LD (r2>0.66) with the top scoring UGT1A1 SNP (rs887829) in European-ancestry populations (Table 2). This is in sharp contrast to the weaker LD (0.26 in HapMap YRI and 0.31 in HUFS) observed between these SNPs in African-ancestry populations (Supplementary Table S3, Supplementary Figure 4). Despite the weaker LD in our African-ancestry population, we were able to directly replicate all three SNPs in African Americans, perhaps indicating the robustness of previous findings.
As described in the materials and methods section, we also conducted local replication analyses of SNPs in LD (r2≥0.3) with the reported SNPs in European- and East-Asian-ancestry populations. This approach was implemented when the reported SNP was not available because it failed one or more of our quality-control filters or was not genotyped or successfully imputed. This approach yielded additional significant associations after Bonferroni correction for multiple testing. For example, SNP rs4148323 in the UGT1A1 gene with a MAF of 0.211 in East Asians was successfully replicated by SNPs in LD with it using the local approach despite been monomorphic in CEU, YRI and African Americans (Table 2, Figure 1). Notably, we successfully replicated all four previously reported UGT1A1 variants from GWAS of serum bilirubin in European- or East-Asian-ancestry populations using either the direct or local replication approaches in our African American study (Table 2).
Corrected P-values and LD in this sample of African Americans (HUFS) for rs4148323 located in an intronic region of the UGT1A1 gene. The red arrow points to the position of the original discovery SNP (rs4148323) in individuals of East Asian ancestry. The two red dots are for SNPs rs887829 and rs6742078 that locally replicated the original discovery SNP (rs4148323) in individuals of East Asian ancestry. Also, SNPs rs887829 and rs6742078 were among the top significant findings in East Asians and Europeans.
In addition, we replicated reported association between variants in the semaphorin 3C (SEMA3C) gene and bilirubin levels.16 The discovery SNP (rs4236644) located in the SEMA3C gene was locally replicated by two LD based SNPs (rs1358503 and rs10251680, P-values 0.0461 and 0.0447, respectively, Figure 2). Each SNP explained ∼1% of the variance in serum bilirubin levels in this study of African Americans.
Corrected P-values and LD in this sample of African Americans (HUFS) for rs4236644 located in the promoter region of the SEMA3C gene. The red arrow points to the position of SNP rs4236644 in Europeans. The two red dots are for SNPs rs1358503 and rs10251660 that locally replicated the original discovery SNP (rs4236644) in individuals of European ancestry. SNPs rs1358503 and rs10251660 are in moderate LD (r2=0.33) with rs4236644 in Europeans.
Discussion
The present study has confirmed the important role of the UGT1A1 gene in serum total bilirubin levels in an African-ancestry population. In this sample of African Americans, the most significantly associated variant (rs887829) is located in the core promoter region of the UGT1A1 gene. Specifically, rs887829 is 211 bp upstream of the short thymine adenine (TA)n repeat sequence covering the TATA box of the UGT1A1 gene. The same SNP was previously reported to be associated with bilirubin levels in populations of East Asians and Europeans.15, 16, 17 Of note, we also replicated the most significant SNP (rs11891311) reported in East Asians and Europeans.15, 16, 17 This SNP is 28 kb upstream from and is in moderate LD (r2=0.31) with rs887829 in African Americans. However, the two SNPs show stronger LD (r2=0.73) in East Asians and Europeans (r2=0.66) as estimated using the HapMap CHB and CEU reference data (Supplementary Table S3 and Supplementary Figure 4). These differences in LD and greater association of rs887829 in HUFS (Table 2) taken together with its functional proximity (ie, near promoter elements) support the likelihood that rs887829 is a more refined proxy for the causative candidate SNP influencing serum total bilirubin levels. Our ability to refine this GWAS signal is because of the lower LD and shorter haplotypes present in African-ancestry populations compared with European- and Asian-ancestry populations; We note that we have successfully refined GWAS signals for fasting plasma glucose and serum uric acid using African ancestry samples in previous studies.27, 32
Interestingly, rs887829 is in strong LD (r2≥0.74) with rs10929302, which was associated with serum total bilirubin with a P-value of 1.37 × 10−11 in the present study. SNP rs10929302 is located in the phenobarbital response enhancer module and known to be associated with irinotecan response.33 This implies that this small (∼3 kb region) UGT1A1 region is pleiotropic, containing variants that influence: (1) serum bilirubin levels, (2) phenobarbital induction of UGT1A1 expression and (3) irinotecan response.
Two families of uridine-diphosphate glucuronosyltransferase (UGT) enzymes are essential to the proper metabolism of a number of exogenous and endogenous compounds. The UGT1 family includes UGT1A1, which has a critical role in the detoxification of neurotoxic bilirubin. Reduced UGT1A1 activity is associated with unconjugated hyperbilirubinemia4 and loss of function has been observed in rare inherited disorders (eg, Gilbert's syndrome and Crigler–Najjar syndrome). Recently, Horsfall et al19 reported that variation in the short (TA)n repeat sequence in the UGT1A1 gene promoter is associated with hyperbilirubinaemia.
There is also considerable continental and ethnic variation in the distribution of the (TA)n repeat in the UGT1A1 gene.19, 34 For example, there are more (TA)n alleles in African populations compared with European-ancestry populations. Alleles (TA)5, (TA)6, (TA)7 and (TA)8 have all been observed in most African populations studied to date; in contrast, alleles (TA)5 and (TA)8 have not been observed in Europeans.19, 34 Also, large difference in allele frequency has been observed across African ethnic groups. The low affinity alleles (TA)7 and (TA)8 are highly prevalent in African populations around the equator.19
Our study emphasizes the importance of conducting GWAS in populations with ancestry from different parts of the world. For example, rs4148323 in UGT1A1 was reported in East Asians15 with a P-value of 1.22 × 10−82. In European (CEU), West African (YRI) and African American (HUFS) samples, this variant is monomorphic compared with a MAF of 0.211 in East Asian (CHB and JPT) HapMap data. However, we were able to replicate this finding by querying a 500 kb-window centered on rs4148323 and performing association analysis on all 61 SNPs in LD using a cutoff r2≥0.3.
GWAS of serum bilirubin in European-ancestry populations also reported significant association with SNP rs4236644 located in the SEMA3C gene; this gene has been linked to increased expression of severe steatotic livers.35 After Bonferroni correction, local replication using rs4236644 as the query SNP, we observed significant association between SNPs (rs1358503 and rs10251680) and bilirubin level (P-values=0.0461 and 0.0447, respectively). In African-ancestry populations, the locally replicated SNPs have higher r2 values with the discovery SNP when compared with European ancestry (Figure 3). Using HapMap CHB data or JPT data, LD is even weaker, which may explain why this SNP was not observed in the GWAS of bilirubin in Koreans.
LD plots from Haploview for a 40-kb region determined by SNP rs4236644 (±20 kb), which located in the promoter region of the SEMA3C gene. Triangle plots were generated from four different HapMap samples of European (CEU), African (YRI), East Asian (CHB) and African American (ASW) ancestries, respectively. Pairwise SNP r2 values are indicated and LD between markers range from complete or strong (black) to weak or no (white) LD. Circled SNPs are rs10251680, rs1358503 and rs4236644.
Despite the small sample size relative to other GWAS, this study had 98 and 94% power, respectively, to replicate the robust effect sizes (1.25 mg/dl and 1.23 mg/dl, respectively), reported for the two top scoring SNPs (rs887829 and rs6742078) in the UGT1A1gene among European and East Asian populations (Table 2).15, 16, 17 However, it had lower power (21%) for a third SNP, rs11891311, reported in an East Asian study.15 Estimated effective sizes in this study of African Americans are 1.25 mg/dl, 1.23 mg/dl and 1.11 mg/dl, respectively, for rs887829, rs6742078 and rs11891311, compared with 1.77 mg/dl, 1.26 mg/dl and 1.2 mg/dl reported in European and East Asian populations.15, 16, 17
In summary, we showed that UGT1A1 is a major locus influencing serum total bilirubin levels in African Americans. Our findings may also contribute to the understanding of the etiology and the treatment of hyperbilirubinaemia in African-ancestry populations. Taking advantage of the lower LD and shorter haplotypes present in African-ancestry populations, we refined previously reported association of rs887829 with serum bilirubin thereby demonstrating the utility of using ethnically diverse populations for replication studies.
References
Arias IM : The excretion of conjugated bilirubin by the liver cell. Medicine (Baltimore) 1966; 45: 513–515.
McDonagh AF : Bilirubin toxicity to human erythrocytes: a more sanguine view. Pediatrics 2007; 120: 175–178.
Ritter JK, Crawford JM, Owens IS : Cloning of two human liver bilirubin UDP-glucuronosyltransferase cDNAs with expression in COS-1 cells. J Biol Chem 1991; 266: 1043–1047.
Pacheco PR, Brilhante MJ, Ballart C et al: UGT1A1, UGT1A6 and UGT1A7 genetic analysis: repercussion for irinotecan pharmacogenetics in the Sao Miguel Island Population (Azores, Portugal). Mol Diagn Ther 2009; 13: 261–268.
Hunt SC, Kronenberg F, Eckfeldt JH, Hopkins PN, Myers RH, Heiss G : Association of plasma bilirubin with coronary heart disease and segregation of bilirubin as a major gene trait: the NHLBI family heart study. Atherosclerosis 2001; 154: 747–754.
Schwertner HA, Jackson WG, Tolan G : Association of low serum concentration of bilirubin with increased risk of coronary artery disease. Clin Chem 1994; 40: 18–23.
Breimer LH, Wannamethee G, Ebrahim S, Shaper AG : Serum bilirubin and risk of ischemic heart disease in middle-aged British men. Clin Chem 1995; 41: 1504–1508.
Lin JP, Vitek L, Schwertner HA : Serum bilirubin and genes controlling bilirubin concentrations as biomarkers for cardiovascular disease. Clin Chem 2010; 56: 1535–1543.
Han SS, Na KY, Chae DW, Kim YS, Kim S, Chin HJ : High serum bilirubin is associated with the reduced risk of diabetes mellitus and diabetic nephropathy. Tohoku J Exp Med 2010; 221: 133–140.
Horsfall LJ, Rait G, Walters K et al: Serum bilirubin and risk of respiratory disease and death. Jama 2011; 305: 691–697.
Han SS, Na KY, Chae DW, Kim YS, Kim S, Chin HJ : Survival benefit of high serum bilirubin and low alkaline phosphatase in older adults. J Am Geriatr Soc 2010; 58: 1413–1415.
Melton PE, Haack K, Goring HH et al: Genetic influences on serum bilirubin in American Indians: The Strong Heart Family Study. Am J Hum Biol 2011; 23: 118–125.
Lin JP, Schwaiger JP, Cupples LA et al: Conditional linkage and genome-wide association studies identify UGT1A1 as a major gene for anti-atherogenic serum bilirubin levels – the Framingham Heart Study. Atherosclerosis 2009; 206: 228–233.
Kronenberg F, Coon H, Gutin A et al: A genome scan for loci influencing anti-atherogenic serum bilirubin levels. Eur J Hum Genet 2002; 10: 539–546.
Kang TW, Kim HJ, Ju H et al: Genome-wide association of serum bilirubin levels in Korean population. Hum Mol Genet 2010; 19: 3672–3678.
Johnson AD, Kavousi M, Smith AV et al: Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet 2009; 18: 2700–2710.
Sanna S, Busonero F, Maschio A et al: Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum Mol Genet 2009; 18: 2711–2718.
Manolio TA, Burke GL, Savage PJ et al: Sex- and race-related differences in liver-associated serum chemistry tests in young adults in the CARDIA study. Clin Chem 1992; 38: 1853–1859.
Horsfall LJ, Zeitlyn D, Tarekegn A et al: Prevalence of clinically relevant UGT1A alleles and haplotypes in African populations. Ann Hum Genet 2011; 75: 236–246.
Adeyemo A, Gerry N, Chen G et al: A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet 2009; 5: e1000564.
Almasy L, Blangero J : Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62: 1198–1211.
McCarroll SA, Kuruvilla FG, Korn JM et al: Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 2008; 40: 1166–1174.
Shriner D, Adeyemo A, Gerry NP et al: Transferability and fine-mapping of genome-wide associated loci for adult height across human populations. PLoS One 2009; 4: e8398.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D : Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Devlin B, Roeder K : Genomic control for association studies. Biometrics 1999; 55: 997–1004.
Ramos E, Chen G, Shriner D et al: Replication of genome-wide association studies (GWAS) loci for fasting plasma glucose in African-Americans. Diabetologia 2011; 54: 783–788.
Bretherton CS WM, Dymnikov VP, Wallace JM, Blade I : The effective number of spatial degrees of freedom of a time-varying field. J Climate 1999; 12: 1990–2009.
Gauderman WJ : Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol 2002; 155: 478–484.
Zucker SD, Horn PS, Sherman KE : Serum bilirubin levels in the U.S. population: gender effect and inverse correlation with colorectal cancer. Hepatology 2004; 40: 827–835.
van Es HH, Bout A, Liu J et al: Assignment of the human UDP glucuronosyltransferase gene (UGT1A1) to chromosome region 2q37. Cytogenet Cell Genet 1993; 63: 114–116.
Charles BA, Shriner D, Doumatey A et al: A genome-wide association study of serum uric acid in African Americans. BMC Med Genomics 2011; 4: 17.
Innocenti F, Kroetz DL, Schuetz E et al: Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics. J Clin Oncol 2009; 27: 2604–2614.
Beutler E, Gelbart T, Demina A : Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci U S A 1998; 95: 8170–8174.
Bertola A, Bonnafous S, Anty R et al: Hepatic expression patterns of inflammatory and immune response genes associated with obesity and NASH in morbidly obese patients. PLoS One 2010; 5: e13577.
Acknowledgements
The study was supported by grants S06GM008016-320107 to CR and S06GM008016-380111 to AA, both from the National Institute of General Medical Sciences MBRS/SCORE Program. Participant enrollment was carried out at the Howard University General Clinical Research Center supported by National Center for Research Resources grant 2M01RR010284 at the National Institutes of Health (NIH). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of NIH or the Department of Health and Human Services. This research was supported in part by the Intramural Research Program of the Center for Research on Genomics and Global Health, which is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the Center for Information Technology and the Office of the Director at the NIH (Z01HG200362). Genotyping support was provided by the Coriell Institute for Medical Research.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Chen, G., Ramos, E., Adeyemo, A. et al. UGT1A1 is a major locus influencing bilirubin levels in African Americans. Eur J Hum Genet 20, 463–468 (2012). https://doi.org/10.1038/ejhg.2011.206
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.206
Keywords
This article is cited by
-
A UGT1A1 variant is associated with serum total bilirubin levels, which are causal for hypertension in African-ancestry individuals
npj Genomic Medicine (2021)
-
The Roles of Common Variation and Somatic Mutation in Cancer Pharmacogenomics
Oncology and Therapy (2019)
-
Serum bilirubin concentration is modified by UGT1A1 Haplotypes and influences risk of Type-2 diabetes in the Norfolk Island genetic isolate
BMC Genetics (2015)
-
Influence of single-nucleotide polymorphisms on deferasirox Ctrough levels and effectiveness
The Pharmacogenomics Journal (2015)
-
Genome-wide analysis of hepatic lipid content in extreme obesity
Acta Diabetologica (2015)