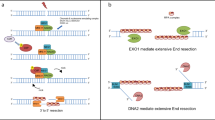
Key Points
-
Ku is a heterodimer composed of two subunits — Ku70 and Ku80 — that binds to DNA ends in a sequence-independent manner. From crystallographic studies, we know that it is shaped like an asymmetric ring, encircling the DNA but also leaving parts of the helix exposed.
-
Ku is found throughout eukaryotic evolution and in higher eukaryotes is associated with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the DNA-PK holoenzyme. Recently, Ku homologues have also been identified in prokaryotes.
-
In all systems studied, Ku has been shown to facilitate the repair of DNA double-strand breaks by non-homologous end-joining (NHEJ).
-
The mechanism by which Ku facilitates NHEJ is not entirely known, but is a consequence of its ability to bind to ends in a sequence-independent manner. For example, Ku might be involved in keeping broken ends together, preventing degradation by nucleases and/or recruiting other DNA-repair factors.
-
Interestingly, Ku has been shown to play a role in numerous other cellular processes, such as immune-system-gene rearrangements, mobile-genetic-element movement, telomere biology, apoptosis and transcription. In many cases, it is clear that the ability to bind to DNA ends in a sequence-independent manner is involved.
Abstract
Ku is of central importance to DNA repair in eukaryotes. In addition, Ku has a key role in a number of other fundamental cellular processes such as telomere maintenance, transcription and apoptosis. The mechanism by which Ku mediates these processes is not entirely understood, but the current knowledge indicates that the function of Ku in these processes might be mechanistically related to its role in DNA repair. Interestingly, recent findings showed that Ku also exists in Archaea and Bacteria, shedding light on aspects of its conservation and evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Mimori, T. et al. Characterization of a high molecular weight acidic nuclear protein recognised by antibodies in sera from patients with polymyositis-scleroderma overlap. J. Clin. Invest. 68, 611–620 (1981).
Mimori, T., Hardin, J. A. & Steitz, J. A. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J. Biol. Chem. 261, 2274–2278 (1986).
Francoeur, A. M., Peebles, C. L., Gompper, P. T. & Tan, E. M. Identification of KI [KU, P70/P80] autoantigens and analysis of anti-KI autoantibody reactivity. J. Immunol. 136, 1648–1653 (1986).
Martin, S. G., Laroche, T., Suka, N., Grunstein, M. & Gasser, S. M. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97, 621–633 (1999). This paper shows that Ku dynamically associates with sites of DNA damage in vivo.
Koike, M., Shiomi, T. & Koike, A. Dimerization and nuclear localization of Ku proteins. J. Biol. Chem. 276, 11167–11173 (2001).
McAinsh, A. D., Scott-Drew, S., Murray, J. A. H. & Jackson, S. P. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr. Biol. 9, 963–966 (1999).
Koike, M. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J. Radiat. Res. 43, 223–236 (2002).
Fewell, J. W. & Kuff, E. L. Intracellular redistribution of Ku immunoreactivity in response to cell–cell contact and growth modulating components in the medium. J. Cell Sci. 109, 1937–1946 (1996).
Sawada, M. et al. Ku70 suppresses the apoptotic translocation of Bax to mitochrondria. Nature Cell Biol. 5, 320–329 (2003).
Morio, T. et al. Ku in the cytoplasm associates with CD40 in human B cells and translocates into the nucleus following incubation with IL-4 and anti-CD40 mAb. Immunity 11, 339–348 (1999).
Gu, Y., Jin, S., Gao, Y., Weaver, D. & Alt, F. W. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl Acad. Sci. USA 94, 8076–8081 (1997).
Singleton, B. K. et al. Molecular and biochemical characterization of xrs mutants defective in Ku80. Mol. Cell. Biol. 17, 1264–1273 (1997).
Errami, A. et al. Ku86 defines the genetic defect and restores X-ray resistance and V(D)J recombination to complementation group 5 hamster cell mutants. Mol. Cell. Biol. 16, 1519–1526 (1996).
Gu, Y. et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7, 653–665 (1997).
Li, G. C. et al. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell 2, 1–8 (1998).
Vogel, H., Lim, D. -S., Karsenty, G., Finegold, M. & Hasty, P. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl Acad. Sci. USA 96, 10770–10775 (1999).
Mimori, T. & Hardin, J. A. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 261, 10375–10379 (1986).
Falzon, M., Fewell, J. W. & Kuff, E. L. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J. Biol. Chem. 268, 10546–10552 (1993).
Yaneva, M., Kowalewski, T. & Lieber, M. R. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 16, 5098–5112 (1997).
Paillard, S., & Strauss, F. Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucl. Acids Res. 19, 5619–5624 (1991).
Ono, M., Tucker, P. W. & Capra, J. D. Production and characterization of recombinant human Ku antigen. Nucl. Acids Res. 22, 3918–3924 (1994).
Pang, D. L., Yoo, S., Dynan, W. S., Jung, M. & Dritschilo, A. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 57, 1412–1415 (1997).
Blier, P. R., Griffith, A. J., Craft, J. & Hardin, J. A. Binding of Ku protein to DNA. J. Biol. Chem. 268, 7594–7601 (1993).
deVries, E., Vandriel, W., Bergsma, W. G., Amberg, A. C. & Vandervliet, P. C. HeLa nuclear-protein recognizing DNA termini and translocating on DNA forming a regular DNA multimeric complex. J. Mol. Biol. 208, 65–78 (1989).
Chiu, C. -F., Lin, T. -Y. & Chou, W. -G. Direct transfer of Ku between DNA molecules with nonhomologous ends. Mutat. Res. 486, 185–194 (2001).
Dynan, W. S. & Yoo, S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucl. Acids Res. 26, 1551–1559 (1998). A comprehensive review of the biochemical properties of Ku and DNA-PK cs.
Gell, D. & Jackson, S. P. Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucl. Acids Res. 27, 3494–3502 (1999).
Aravind, L. & Koonin, E. V. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 11, 1365–1374 (2001). This report, in addition to reference 46, identified the presence of Ku homologues in prokaryotes.
Aravind, L. & Koonin, E. V. SAP — a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25, 112–114 (2000).
Walker, J. R., Corpina, R. A. & Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 (2001). Report of the crystal structure of Ku alone and bound to DNA, which provided enormous insight into the biochemical properties of Ku including the mechanism of DNA binding.
Wang, J., Satoh, M., Chou, C. & Reeves, W. H. Similar DNA binding properties of free p70 (KU) subunit and p70/p80 heterodimer. FEBS Lets. 351, 219–224 (1994).
Griffith, A. J., Blier, P. R., Mimori, T. & Hardin, J. A. Ku polypeptides synthesized in vitro assemble into complexes which recognize ends of double-stranded DNA. J. Biol. Chem. 267, 331–338 (1992).
Zhang, Z. et al. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J. Biol. Chem. 276, 38231–38236 (2001).
Harris, R. et al. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR). J. Mol. Biol. 335, 573–582 (2004).
Giffen, W. et al. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature 380, 265–268 (1996).
Schild-Poulter, C. et al. Differential DNA binding of Ku antigen determines its involvement in DNA replication. DNA Cell Biol. 22, 65–78 (2003).
Gottlieb, T. M. & Jackson, S. P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72, 131–142 (1993).
Dvir, A., Peterson, S. R., Knuth, M. W., Lu, H. & Dynan, W. S. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl Acad. Sci. USA 89, 11920–11924 (1992). References 37 and 38 show that Ku is a part of the DNA-dependent protein kinase in higher eukaryotes.
Suwa, A. et al. DNA-dependent protein kinase (Ku protein–p350 complex) assembles on double-stranded DNA. Proc. Natl Acad. Sci. USA 91, 6904–6908 (1994).
Hammarsten, O. & Chu, G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc. Natl Acad. Sci. USA 95, 525–530 (1998).
Chan, D. W. & Lees-Miller, S. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 271, 8936–8941 (1996).
Myung, K., He, D. M., Lee, S. E. & Hendrickson, E. A. KARP-1: a novel leucine zipper protein expressed from the Ku86 autoantigen locus is implicated in the control of DNA-dependent protein kinase activity. EMBO J. 18, 3172–3184 (1997).
Hanakahi, L. A. & West, S. C. Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK. EMBO J. 21, 2038–2044 (2002).
Ma, Y. & Lieber, M. R. Binding of inositol hexakisphosphate (IP6) to Ku but not to DNA-PKcs. J. Biol. Chem. 277, 10756–10759 (2002).
Hanakahi, L. A., Bartlet-Jones, M., Chappell, C., Pappin, D. & West, S. C. Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell 102, 721–729 (2000).
Doherty, A. J., Jackson, S. P. & Weller, G. R. Identification of bacterial homologes of the Ku DNA-repair proteins. FEBS Lett. 500, 186–188 (2001).
West, S. C. Molecular views of recombination proteins and their control. Nature Rev. Mol. Cell Biol. 4, 1–11 (2003).
Lieber, M. R., Ma, Y., Pannicke, U. & Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nature Rev. Mol. Cell Biol. 4, 712–720 (2003). A comprehensive review of NHEJ.
Jeggo, P. A., Taccioli, G. E. & Jackson, S. P. Menage a trois: double strand break repair, V(D)J recombination and DNA-PK. Bioessays 17, 949–957 (1995).
Ferguson, D. O. & Alt, F. W. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene 20, 5572–5579 (2001).
Smith, G. C. M. & Jackson, S. P. The DNA-dependent protein kinase. Genes Dev. 13, 916–934 (1999).
Baumann, P. & West, S. C. DNA end-joining cayalyzed by human cell free extracts. Proc. Natl Acad. Sci. USA 95, 14066–14070 (1998).
Labhart, P. Nonhomologous DNA end joining in cell-free systems. Eur. J. Biochem. 265, 849–861 (1999).
Ramsden, D. A. & Gellert, M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 17, 609–614 (1998).
McElhinny, S. A. N., Snowden, C. M., McCarville, J. & Ramsden, D. A. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20, 2996–3003 (2000).
Chen, L., Trujillo, K., Sung, P. & Tomkinson, A. E. Interactions of the DNA ligase IV–XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem. 275, 26196–26205 (2000).
Feldmann, H. & Winnacker, E. L. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J. Biol. Chem. 268, 12895–12900 (1993).
Siede, W., Friedl, A. A., Dianova, I., Eckardt-Schupp, F. & Friedberg, E. C. The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics 142, 91–102 (1996).
Boulton, S. J. & Jackson, S. P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15, 5093–5103 (1996).
Tsukamoto, Y., Kato, J. & Ikeda, H. Hdf1, a yeast Ku protein homologue, is involved in illegitimate recombination, but not in homologous recombination. Nucl. Acids Res. 24, 2067–2072 (1996).
Milne, G. T., Jin, S., Shannon, K. B. & Weaver, D. T. Mutations in two Ku homologues define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 4189–4198 (1996). References 57–61 were instrumental in demonstrating Ku-dependent NHEJ in S. cerevisiae.
Barnes, G. & Rio, D. DNA double-strand break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 94, 867–872 (1997).
Chen, L., Trujillo, K., Ramos, W., Sung, P. & Tomkinson, A. E. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8, 1105–1115 (2001).
West, C. E. et al. Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 31, 517–528 (2002).
Gallego, M. E., Bleuyard, J. -Y., Daoudal-Cotterell, S., Jallut, N. & White, C. I. Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 35, 557–565 (2003).
Kooistra, R., Pastink, A., Zonneveld, J. B. M., Lohman, P. H. M. & Eeken, J. C. J. The Drosophila melanogaster DmRAD54 gene plays a crucial role in double-strand break repair after P-element excision and acts synergistically with Ku70 in the repair of X-ray damage. Mol. Cell. Biol. 19, 6269–6275 (1999).
Conway, C. et al. Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J. Biol. Chem. 277, 21269–21277 (2002).
Weller, G. R. et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297, 1686–1689 (2002). Shows that the putative Ku homologues from prokaryotes are functional homologues and are likely to have a role in NHEJ in bacteria.
Lee, S. E. et al. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 399–409 (1998).
Liang, F. & Jasin, M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 271, 14405–14411 (1996).
Hsu, H. -L., Yannone, S. M. & Chen, D. J. Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair 1, 225–235 (2002).
Karmakar, P., Snowden, C. M., Ramsden, D. A. & Bohr, V. A. Ku heterodimer binds to both ends of the Werner protein and functional interaction occurs at the Werner N-terminus. Nucl. Acids Res. 30, 3583–3591 (2002).
Cooper, M. P. et al. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 14, 907–912 (2000).
Goedecke, W., Eijpe, M., Offenberg, H. H., van Aalderen, M. & Heyting, C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nature Genet. 23, 194–198 (1999).
Galande, S. & Kohwi-Shigematsu, T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274, 20521–20528 (1999).
Van Dyck, E., Stasiak, A. Z., Stasiak, A. & West, S. C. Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398, 728–731 (1999).
Pierce, A. J., Hu, P., Han, M., Ellis, N. & Jasin, M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15, 3237–3242 (2001).
Allen, C., Kurimasa, A., Brenneman, M. A., Chen, D. J. & Nickoloff, J. A. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl Acad. Sci. USA 99, 3758–3763 (2002).
Ristic, D., Modesti, M., Kanaar, R. & Wyman, C. Rad52 and Ku bind to different DNA structures produced early in double-strand break repair. Nucl. Acids Res. 31, 5229–5237 (2003).
Rodgers, W., Jordan, S. J. & Capra, J. D. Transient association of Ku with nuclear substrates characterized using fluorescence photobleaching. J. Immunol. 168, 2348–2355 (2002).
Yavuzer, U., Smith, G. C. M., Bliss, T., Werner, D. & Jackson, S. P. DNA end-independent activation of DNA-PK mediated via association with the DNA-binding protein C1D. Genes Dev. 12, 2188–2199 (1998).
Wang, X., Li, G. C., Iliakis, G. & Wang, Y. Ku affects the CHK1-dependent G2 checkpoint after ionizing radiation. Cancer Res. 62, 6031–6034 (2002).
Zhou, X. -Y. et al. Ku affects the ATM-dependent S phase checkpoint following ionizing radiation. Oncogene 21, 6377–6381 (2002).
Casellas, R. et al. Ku80 is required for immunoglobulin isotype switching. EMBO J. 17, 2404–2411 (1998).
Manis, J. P. et al. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 187, 2081–2089 (1998).
Taccioli, G. E. et al. Ku80: Product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 265, 1442–1445 (1994).
Bassing, C. H., Swat, W. & Alt, F. W. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109, S45–S55 (2002).
Zhu, C., Bogue, M. A., Lim, D., Hasty, P. & Roth, D. B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86, 379–389 (1996).
Kulesza, P. & Lieber, M. R. DNA-PK is essential only for coding joint formation in V(D)J recombination. Nucl. Acids Res. 26, 3944–3948 (1998).
Nussenzweig, A. et al. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382, 551–555 (1996).
Barnes, D. E., Stamp, G., Rosewell, I., Denzel, A. & Lindahl, T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8, 1395–1398 (1998).
Han, J., Steen, S. B. & Roth, D. B. Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol. Cell. Biol. 17, 2226–2234 (1997).
Purugganan, M. M., Shah, S., Kearney, J. F. & Roth, D. B. Ku80 is required for addition of N nucleotides to V(D)J recombination junctions by terminal deoxynucleotidyl transferase. Nucl. Acids Res. 29, 1638–1646 (2001).
Mills, K. D., Ferguson, D. O. & Alt, F. W. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194, 77–95 (2003).
Difilippantonio, M. J. et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404, 510–514 (2000). The first demonstration that Ku functions to prevent tumourigenesis in mammals.
Downs, J. A. & Jackson, S. P. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol. Cell. Biol. 19, 6260–6268 (1999).
Daniel, R., Katz, R. A. & Skalka, A. M. A role for DNA-PK in retroviral DNA integration. Science 284, 644–647 (1999).
Li, L. et al. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20, 3272–3281 (2001). References 96, 97 and 98 show the involvement of Ku in retroelement integration, in which there is no DNA DSB created in the host genome.
d'Adda di Fagagna, F., Weller, G. R., Doherty, A. J. & Jackson, S. P. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 4, 47–52 (2003).
Akroyd, J. & Symonds, N. Localization of the gam gene of bacteriophage Mu and characterisation of the gene product. Gene 49, 273–282 (1986).
Abraham, Z. H. L. & Symonds, N. Purification of overexpressed gam gene protein from bacteriophage Mu by denaturation-renaturation techniques and a study of its DNA-binding properties. Biochem. J. 269, 679–684 (1990).
Hediger, F., Neumann, F. R., Van Houwe, G., Dubrana, K. & Gasser, S. M. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways. Curr. Biol. 12, 2076–2089 (2002).
Nugent, C. I. et al. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8, 657–660 (1998).
Boulton, S. J. & Jackson, S. P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17, 1819–1828 (1998).
Gravel, S., Larrivee, M., Labrecque, P. & Wellinger, R. J. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280, 741–744 (1998).
Porter, S. E., Greenwell, P. W., Ritchie, K. B. & Petes, T. D. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucl. Acids Res. 24, 582–585 (1996).
Gravel, S. & Wellinger, R. J. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol. Cell. Biol. 22, 2182–2193 (2002).
Cosgrove, A. J., Nieduszynski, C. A. & Donaldson, A. D. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 16, 2485–2490 (2002).
Bertuch, A. A. & Lundblad, V. The Ku heterodimer performs separable activities at double strand breaks and chromosome termini. Mol. Cell. Biol. 23, 8202–8215 (2003).
Stellwagen, A. E., Haimberger, Z. W., Veatch, J. R. & Gottschling, D. E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17, 2384–2395 (2003).
Roy, R., Meier, B., McAinsh, A. D., Feldmann, H. M. & Jackson, S. P. Separation-of-function mutants of yeast Ku80 reveal a Yku80p–Sir4p interaction involved in telomeric silencing. J. Biol. Chem. 279, 86–94 (2004). References 109, 110 and 111 report the identification of separation-of-function mutations in Ku80, which show separate roles for Ku in NHEJ and at telomeres.
Baumann, P. & Cech, T. R. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell 11, 3265–3275 (2000).
Riha, K., Watson, J. M., Parkey, J. & Shippen, D. E. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21, 2819–2826 (2002).
Bundock, P., van Attikum, H. & Hooykaas, P. Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucl. Acids Res. 30, 3395–3400 (2002).
Miyoshi, T., Sadaie, M., Kanoh, J. & Ishikawa, F. Telomeric DNA ends are essential for the localization of Ku at telomeres in fission yeast. J. Biol. Chem. 278, 1924–1931 (2003).
Hsu, H., Gilley, D., Blackburn, E. H. & Chen, D. J. Ku is associated with the telomere in mammals. Proc. Natl Acad. Sci. USA 96, 12454–12458 (1999).
d'Adda di Fagagna, F. et al. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11, 1192–1196 (2001).
Chai, W., Ford, L. P., Lenertz, L., Wright, W. E. & Shay, J. W. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J. Biol. Chem. 277, 47242–47247 (2002).
Espejel, S. et al. Mammalian Ku86 mediates chromosome fusions and apoptosis caused by critically short telomeres. EMBO J. 21, 2207–2219 (2002).
Smogorzewska, A., Karlseder, J., Holtgreve-Grez, H., Jauch, A. & de Lange, T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12, 1635–1644 (2002).
Mo, X. & Dynan, W. S. Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Mol. Cell. Biol. 22, 8088–8099 (2002).
Woodard, R. L., Lee, K., Huang, J. & Dynan, W. S. Distinct roles for Ku protein in transcriptional reinitiation and DNA repair. J. Biol. Chem. 276, 15423–15433 (2001).
Kuhn, A., Gottleib, T. M., Jackson, S. P. & Grummt, I. DNA-dependent protein kinase: a potent inhibitor of transcription by RNA polymerase I. Genes Dev. 9, 193–203 (1995).
Manis, J., Tian, M. & Alt, F. W. Mechanism and control of class-switch recombination. Trends Immunol. 23, 31–39 (2002).
Neuberger, M. S., Harris, R. S., Di Noia, J. & Petersen-Mahrt, S. K. Immunity through DNA deamination. Trends Biochem. Sci. 28, 305–312 (2003).
Acknowledgements
We are grateful to A. Harvey and S. Bell for help with the figures.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- AUTOANTIGEN
-
An endogenous antigen that stimulates the production of autoantibodies.
- AUTOANTIBODY
-
An antibody that reacts with the cells or tissues of the organism in which it was produced.
- CLASS-SWITCH RECOMBINATION
-
After immune cells have undergone V(D)J recombination, they can change the constant region of their antibodies in response to stimulation by antigen. This results in changes in the effector functions of the antibodies. The process is initiated by the generation of double-stranded breaks in a region-specific manner, and some components of the non-homologous end-joining machinery, including Ku, have been implicated in their subsequent repair.
- DNaseI FOOTPRINTING
-
An in vitro method for determining the location and manner of protein binding to DNA by examining the pattern of DNaseI-mediated DNA degradation with and without the protein of interest.
- ELECTROPHORETIC MOBILITY SHIFT ASSAY
-
(EMSA). An in vitro method for detecting protein–DNA interactions by analyzing the migration on a native polyacrylamide gel of radiolabelled DNA with and without the protein of interest.
- INOSITOL HEXAKISPHOSPHATE
-
(IP6). A phospholipid that is involved in intracellular signalling.
- HOMOLOGOUS RECOMBINATION
-
A DNA-recombination pathway, which includes the repair of double-stranded DNA breaks, that uses a homologous double-stranded DNA molecule as a template for the repair of the broken DNA.
- MRN COMPLEX
-
In mammalian cells, this complex is made up of MRE11, RAD50 and NBS1. In S. cerevisiae, it is made up of the proteins Mre11, Rad50 and Xrs2 (hence the name MRX). Mre11 has been shown to have nuclease activity, and it is thought that this complex has a role in both homologous recombination and non-homologous end-joining.
- CHROMATIN IMMUNOPRECIPITATION
-
(ChIP). A method for determining whether a protein binds to a particular region of the genome in vivo. It involves treating live cells with formaldehyde to form nonspecific crosslinks between the DNA and associated proteins. The cells are then lysed, the genomic DNA is sheared into small fragments and the protein of interest is immunoprecipitated. Any protein-associated DNA is then removed and analysed by quantitative PCR.
- EXONUCLEASE
-
An enzyme that degrades nucleic acids in a manner that requires an available end.
- FLUORESCENCE PHOTOBLEACHING
-
A method for studying the dynamics of proteins in vivo. The protein of interest is fused to a moiety that is able to fluoresce, and this fluorescence can be altered by treating cells with a laser pulse (photobleaching), thereby allowing the study of protein mobility in the cell by examining the changes in fluorescence over time after the application of the laser pulse.
- SOMATIC HYPERMUTATION
-
A mechanism for creating extra variability in antibody genes that occurs after V(D)J recombination, by introducing point mutations, small insertions and small deletions into the V(D)J coding sequences.
- ENDONUCLEASE
-
An enzyme that catalyses the hydrolytic cleavage of DNA in the middle of a DNA strand or double helix.
- TRANSPOSON
-
A mobile genetic element that can relocate within the genome of their hosts. An autonomous transposon encodes a transposase protein that catalyses its excision and reintegration in the genome, and can therefore direct its own transposition.
- RETROTRANSPOSON
-
A mobile genetic element that is first transcribed and then reverse-transcribed and the cDNA is inserted into the host genome. In the case of autonomous retrotransposons, the reverse transcription and integration are performed by retrotransposon-encoded reverse transcriptase and integrase proteins, respectively.
- RETROVIRUS
-
An RNA virus that encodes an RNA-dependent DNA polymerase, known as reverse transcriptase, and behaves as a retrotransposon.
- TELOMERASE
-
An enzyme that is capable of extending the ends of telomeres after replication by using an RNA template that is part of the enzyme complex.
Rights and permissions
About this article
Cite this article
Downs, J., Jackson, S. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol 5, 367–378 (2004). https://doi.org/10.1038/nrm1367
Issue Date:
DOI: https://doi.org/10.1038/nrm1367
This article is cited by
-
Molecular cloning, subcellular localization, and rapid recruitment to DNA damage sites of chicken Ku70
Scientific Reports (2024)
-
CRISPR/Cas9 system is a suitable gene targeting editing tool to filamentous fungus Monascus pilosus
Applied Microbiology and Biotechnology (2024)
-
DNA damage repair and cancer immunotherapy
Genome Instability & Disease (2023)
-
Cancer-associated fibroblast-induced lncRNA UPK1A-AS1 confers platinum resistance in pancreatic cancer via efficient double-strand break repair
Oncogene (2022)
-
CRL4DCAF8 dependent opposing stability control over the chromatin remodeler LSH orchestrates epigenetic dynamics in ferroptosis
Cell Death & Differentiation (2021)