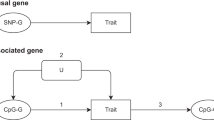
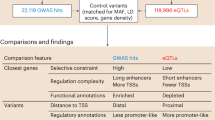
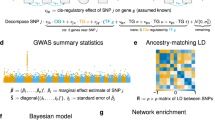
Abstract
A key goal of biomedical research is to elucidate the complex network of gene interactions underlying complex traits such as common human diseases. Here we detail a multistep procedure for identifying potential key drivers of complex traits that integrates DNA-variation and gene-expression data with other complex trait data in segregating mouse populations. Ordering gene expression traits relative to one another and relative to other complex traits is achieved by systematically testing whether variations in DNA that lead to variations in relative transcript abundances statistically support an independent, causative or reactive function relative to the complex traits under consideration. We show that this approach can predict transcriptional responses to single gene–perturbation experiments using gene-expression data in the context of a segregating mouse population. We also demonstrate the utility of this approach by identifying and experimentally validating the involvement of three new genes in susceptibility to obesity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Hughes, T.R. et al. Functional discovery via a compendium of expression profiles. Cell 102, 109–126 (2000).
Karp, C.L. et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat. Immunol. 1, 221–226 (2000).
Schadt, E.E. et al. Genetics of gene expression surveyed in maize, mouse and man. Nature 422, 297–302 (2003).
Johnson, J.M. et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144 (2003).
Schadt, E.E. A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 5, R73 (2004).
Shoemaker, D.D. et al. Experimental annotation of the human genome using microarray technology. Nature 409, 922–927 (2001).
DePrimo, S.E. et al. Expression profiling of blood samples from an SU5416 Phase III metastatic colorectal cancer clinical trial: a novel strategy for biomarker identification. BMC Cancer 3, 3 (2003).
Mootha, V.K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
van't Veer, L.J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
Waring, J.F. et al. Identifying toxic mechanisms using DNA microarrays: evidence that an experimental inhibitor of cell adhesion molecule expression signals through the aryl hydrocarbon nuclear receptor. Toxicology 181–182, 537–550 (2002).
Monks, S.A. et al. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 75, 1094–1105 (2004).
Morley, M. et al. Genetic analysis of genome-wide variation in human gene expression. Nature 430, 743–747 (2004).
Zhu, J. et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet. Genome Res. 105, 363–374 (2004).
Klose, J. et al. Genetic analysis of the mouse brain proteome. Nat. Genet. 30, 385–393 (2002).
Luscombe, N.M. et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431, 308–312 (2004).
Brem, R.B., Yvert, G., Clinton, R. & Kruglyak, L. Genetic dissection of transcriptional regulation in budding yeast. Science 296, 752–755 (2002).
Yvert, G. et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35, 57–64 (2003).
Pearl, J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference (Morgan Kaufmann, San Mateo, California, 1988).
Sakamoto, Y., Ishiguro, M. & Kitagawa, G. Akaike Information Criterion Statistics (D. Reidel, Dordrecht, The Netherlands, 1986).
Jiang, C. & Zeng, Z.B. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140, 1111–1127 (1995).
Drake, T.A. et al. Genetic loci determining bone density in mice with diet-induced atherosclerosis. Physiol. Genomics 5, 205–215 (2001).
Laurie, C.C. et al. The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics 168, 2141–2155 (2004).
Zeng, Z.B. et al. Genetic architecture of a morphological shape difference between two Drosophila species. Genetics 154, 299–310 (2000).
Chesler, E.J. et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37, 233–242 (2005).
Ghazalpour, A. et al. Genomic analysis of metabolic pathway gene expression associated with obesity. Genome Biol. (in the press).
Grant, G.R., Liu, J. & Stoeckert, C.J., Jr. A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics 21, 2684–2690 (2005).
Masuzaki, H. et al. A transgenic model of visceral obesity and the metabolic syndrome. Science 294, 2166–2170 (2001).
Rask, E. et al. Tissue-specific changes in peripheral cortisol metabolism in obese women: increased adipose 11beta-hydroxysteroid dehydrogenase type 1 activity. J. Clin. Endocrinol. Metab. 87, 3330–3336 (2002).
Alberts, P. et al. Selective inhibition of 11beta-hydroxysteroid dehydrogenase type 1 decreases blood glucose concentrations in hyperglycaemic mice. Diabetologia 45, 1528–1532 (2002).
Alberts, P. et al. Selective inhibition of 11 beta-hydroxysteroid dehydrogenase type 1 improves hepatic insulin sensitivity in hyperglycemic mice strains. Endocrinology 144, 4755–4762 (2003).
Alessi, M.C. et al. Plasminogen activator inhibitor 1, transforming growth factor-beta1, and BMI are closely associated in human adipose tissue during morbid obesity. Diabetes 49, 1374–1380 (2000).
Romano, M. et al. Association of inflammation markers with impaired insulin sensitivity and coagulative activation in obese healthy women. J. Clin. Endocrinol. Metab. 88, 5321–5326 (2003).
Rosmond, R., Chagnon, M., Bouchard, C. & Bjorntorp, P. Increased abdominal obesity, insulin and glucose levels in nondiabetic subjects with a T29C polymorphism of the transforming growth factor-beta1 gene. Horm. Res. 59, 191–194 (2003).
Samad, T.A., Krezel, W., Chambon, P. & Borrelli, E. Regulation of dopaminergic pathways by retinoids: activation of the D2 receptor promoter by members of the retinoic acid receptor-retinoid X receptor family. Proc. Natl. Acad. Sci. USA 94, 14349–14354 (1997).
Schupf, N., Williams, C.A., Hugli, T.E. & Cox, J. Psychopharmacological activity of anaphylatoxin C3a in rat hypothalamus. J. Neuroimmunol. 5, 305–316 (1983).
Choy, L.N. & Spiegelman, B.M. Regulation of alternative pathway activation and C3a production by adipose cells. Obes. Res. 4, 521–532 (1996).
Pomeroy, C. et al. Effect of body weight and caloric restriction on serum complement proteins, including Factor D/adipsin: studies in anorexia nervosa and obesity. Clin. Exp. Immunol. 108, 507–515 (1997).
Ylitalo, K. et al. Serum complement and familial combined hyperlipidemia. Atherosclerosis 129, 271–277 (1997).
Lange, R. et al. Developmentally regulated mouse gene NK10 encodes a zinc finger repressor protein with differential DNA-binding domains. DNA Cell Biol. 14, 971–981 (1995).
Goodarzi, M.O. et al. Lipoprotein lipase is a gene for insulin resistance in Mexican Americans. Diabetes 53, 214–220 (2004).
Carlborg, O. et al. Methodological aspects of the genetic dissection of gene expression. Bioinformatics 21, 2383–2393 (2005).
Kao, C.H. & Zeng, Z.B. Modeling epistasis of quantitative trait loci using Cockerham's model. Genetics 160, 1243–1261 (2002).
Sillanpaa, M.J. & Corander, J. Model choice in gene mapping: what and why. Trends Genet. 18, 301–307 (2002).
He, Y.D. et al. Microarray standard data set and figures of merit for comparing data processing methods and experiment designs. Bioinformatics 19, 956–965 (2003).
Hughes, T.R. et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19, 342–347 (2001).
Jiang, C. & Zeng, Z.B. Mapping quantitative trait loci with dominant and missing markers in various crosses from two inbred lines. Genetica 101, 47–58 (1997).
Haley, C.S. & Knott, S.A. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69, 315–324 (1992).
Miller, A.J. Subset Selection in Regression (Chapman and Hall, London; New York, 1990).
Broman, K.W. PhD Dissertation: Identifying Quantitative Trait Loci in Experimental Crosses (University of California, Berkeley, 1997).
Acknowledgements
This work was supported in part by grants from the US National Institutes of Health (A.J.L.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Testing the power of the LCMS procedure to identify relationships among complex traits. (PDF 26 kb)
Supplementary Fig. 2
Disruption of C3ar1. (PDF 34 kb)
Supplementary Fig. 3
Diagram outlining the multi-step procedure defined in the main text to identify causal genes for obesity in mice. (PDF 14 kb)
Supplementary Fig. 4
Disruption of Tgfbr2. (PDF 30 kb)
Supplementary Fig. 5
Expression of the human ZFP90 transgene and the murine Zfp90 gene in 6 mouse tissues. (PDF 52 kb)
Supplementary Table 1
Liver gene expression traits significantly correlated with omental fat pad mass in the BXD cross. (PDF 24 kb)
Supplementary Table 2
Genes with at least 2 eQTL overlapping OFPM QTL tested for causal associations to OFPM. (PDF 8 kb)
Rights and permissions
About this article
Cite this article
Schadt, E., Lamb, J., Yang, X. et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37, 710–717 (2005). https://doi.org/10.1038/ng1589
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1589
This article is cited by
-
A novel approach for predicting upstream regulators (PURE) that affect gene expression
Scientific Reports (2023)
-
Leveraging molecular quantitative trait loci to comprehend complex diseases/traits from the omics perspective
Human Genetics (2023)
-
Genetic analysis of blood molecular phenotypes reveals common properties in the regulatory networks affecting complex traits
Nature Communications (2023)
-
DrGA: cancer driver gene analysis in a simpler manner
BMC Bioinformatics (2022)
-
Traversing industry and academia in biomedicine: the best of both worlds?
Nature Reviews Genetics (2022)