Article Text
Abstract
Background The majority of lung cancers are smoking-related, with environmental and genetical factors contributing. The interplay between environmental and genetical contributions in non-smoking-related lung cancers is less clear.
Methods We analysed a population-based computerised genealogy resource linked to a state-wide cancer registry of lung cancer cases (n=5544) for evidence of a genetical contribution to lung cancer predisposition in smoking (n=1747) and non-smoking cases (n=784). Statistical methods were used to test for significant excess relatedness of cases and estimate relative risk (RR) in close and distant relatives of lung cancer cases.
Results Significant excess relatedness was observed for all lung cancer cases (p<0.001) and for the subsets of smoking-related (p<0.001) and non-smoking-related (p<0.001) cases when all pairwise relationships were considered. Only the non-smoking-related subset of cases showed significant excess relatedness when close relationships were ignored (p=0.020). First-degree, second-degree, and fourth-degree relatives of non-smoking-related lung cancer cases had significantly elevated RR. An even higher elevated RR was observed for first-degree, second-degree, third-degree and fourth-degree relatives of smoking-related lung cancer cases.
Conclusions Non-smoking-related lung cancer cases show significant excess relatedness for close and distant relationships, providing strong evidence for a genetical contribution as well as an environmental contribution. Significant excess relatedness for only close family relationships in all lung cancer cases and in only smoking-related lung cancer cases implies environmental contribution. Additionally, the highest RR for lung cancer was observed in the relatives of smoking-related lung cancer, suggesting predisposition gene carriers who smoke are at highest risk for lung cancer. Screening and gene identification should focus on high-risk pedigrees.
- Lung Cancer
- Clinical Epidemiology
Statistics from Altmetric.com
Key messages
What is the key question?
How much does genetics contribute to smoking-related and non-smoking-related lung cancer?
What is the bottom line?
There is strong evidence for a genetical contribution in non-smoking-related lung cancer out to fourth degree relatives.
Why read on?
The relative risk of developing lung cancer in smokers and non-smokers with a family history of lung cancer varies based upon genetical distance.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide.1 In USA it was estimated to account for 159 260 deaths in 2014.1 ,2 Overall survival from lung cancer at 5 years remains below 16% and has not changed in the past 30 years. Recently, low-dose screening for high-risk individuals has been shown to result in a 20% relative risk (RR) reduction in mortality from lung cancer.3 Follow-up studies support this conclusion, lending support to lung cancer screening efforts. The major variables that define high-risk individuals are smoking and age.
While cigarette smoking is well established as the strongest risk factor for the development of lung cancer, environmental and genetical factors are suspected to play a role.4 ,5 Studies have shown familial clustering of lung cancer cases, supporting evidence for a genetical contribution to the development of lung cancer.6–12 These show an approximate twofold increase in lung cancer development in first-degree relatives. Similar results are observed in the Utah population.11 ,12 This has led some to incorporate family history into lung cancer risk models.13
Presence of familial clustering in close relatives is supportive, but not sufficient, to prove a genetical contribution; the aetiology may be partly explained by shared environment and risk factors in close relatives, but may also include shared genetical predisposition. Non-genetical environmental exposures such as primary cigarette smoking, secondhand smoke, or shared occupations or behaviours can confound any evidence of genetical contribution to lung cancer, especially in close relatives.14–17 Consequently, it remains difficult to show a clear genetical contribution to the risk of developing lung cancer. Evaluation of increased risk beyond first-degree relationships would provide greater weight to a genetical contribution; evaluation of familial clustering and risk in the absence of smoking would eliminate the largest confounding variable.
The unique Utah Population Database (UPDB) links Utah Cancer Registry (UCR) data from 1966 with genealogy data representing the state of Utah from the mid-1800s, and with Utah death certificate (DC) data from 1904. This allowed us to analyse and examine evidence for a significant heritable contribution to predisposition to lung cancer in non-smoking lung cancer cases.
Data and methods
Utah population database
Originally created in the 1970s using family history data from the Family History Library of the Church of Jesus Christ of Latter-day Saints (Mormons), and since expanded using Utah vital statistics data, UPDB is a computerised genealogy of the Utah pioneers and their descendants.18 The original genealogy included 1.6 million individuals and extended to six generations, it currently has over 7 million unique individuals; some pedigrees extend to 12 generations. A smaller set of 1.35 million individuals in the UPDB has at least 12 of their 14 immediate ancestors (both parents, all 4 grandparents, and at least 6 of their 8 great grandparents) in the genealogy. Individuals meeting these strict criteria were considered to have good genealogy content and were used for the genetical analyses to ensure individuals analysed had similar data available for relatives. All such individuals in the UPDB were assigned to cohorts based on 5-year birth year, sex and place of birth (Utah or not). This allowed selection of matched controls for cases, and allowed estimation of cohort-specific rates of lung cancer. Genealogy data in the UPDB has been record linked to various databases, including the UCR and the computerised Utah DCs.
Utah Cancer Registry
The UCR, a founding member of the National Cancer Institute’s SEER Program, has continuously collected data on every cancer occurring in the State of Utah since 1966. Only independent primary cancers are recorded in the UCR; cancer includes all in situ (except in situ cervical cancers) or malignant neoplasms (excluding basal and squamous cell carcinomas of the skin except in genital sites); over 98% follow-up is achieved.
Lung cancer cases were identified in the UCR based on primary site coding of the International Classification of Diseases for Oncology (C340–349; bronchus and lung), including histology codes 8000–9589 (which excludes leukaemias and lymphomas) in the UCR.
Utah DCs: smoking status
Utah DCs from 1904 are computerised and record linked to the Utah genealogy data. Deaths from 1904 to 1956 were coded to International Classification of Disease revision 10; other deaths are coded to International Classification of Disease revisions 6–10, according to the year of death. All Utah DCs include primary cause of death, and many include contributing causes of death.
There are six separate distinctions for the tobacco contribution to death on a Utah DC. The category ‘Non-user’ identified non-smokers. Two categories, ‘Was underlying cause of death’ and ‘Probably contributed to the cause of death’ were used to identify smokers. The categories ‘Did not contribute to cause of death’, ‘Unknown in relation to cause of death’, and ‘Unknown if user’ were excluded from analysis.
Genealogical index of familiality test for excess relatedness
The genealogical index of familiality (GIF) test for excess relatedness was designed specifically for use with the Utah genealogy and considers the average pairwise relatedness for a set of individuals.12 The test is accomplished by comparing the average pairwise relatedness of all pairs of individuals in a set (eg, all lung cancer cases) with their expected average pairwise relatedness in the Utah population, as estimated in 1000 sets of matched controls from the UPDB. The pairwise relatedness for a pair is measured as the Malécot's coefficient of kinship,19 which measures the probability that randomly selected homologous genes from a common ancestor are shared identical by descent. The coefficient is based on the genetical distance between the pairs, through all common ancestors. The value is 1/2 for parent/offspring, 1/4 for siblings or grandparent/grandchild, and 1/8 for avunculars, for example. Unrelated individuals have a coefficient of 0. The GIF is multiplied by 105 for ease of presentation.
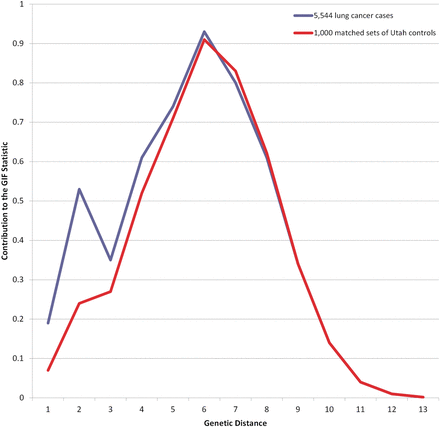
To calculate the significance of the GIF test we select 1000 sets of matched (5-year birth year, sex and place of birth) controls from the UPDB and calculate 1000 GIF statistics for these sets. The average pairwise relatedness for the cases is compared with the distribution of the 1000 control GIFs to ascertain the empirical significance of the GIF test. Matched controls for each lung cancer case and smoking subsets were selected from all individuals in the UPDB with good genealogy content. A GIF figure shows the contribution to the mean case GIF statistic (y axis) by the genetical distance of the pair (x axis).
The overall GIF test performs a test of the hypothesis of no excess familial clustering; the test considers all pairwise relationships. A disease with a strong environmental factor and no genetical contribution might show excess familial clustering, but this would only be exhibited in close relationships, for which individuals share environment and behaviour. For this reason we also consider the distant GIF (dGIF) test, which is similar to the overall GIF test, but ignores all pairwise relationships closer than first cousins (genetical distance 4), in cases and controls. A significant dGIF statistic shows evidence of excess relatedness among distant relationships, and provides strong support for a genetical contribution to the phenotype tested.
Relative risks in relatives
Estimation of RR as the ratio of the observed number of affected relatives among a set of relatives to the expected number of affected relatives is the most common method to evaluate genetical contribution to phenotype. Cohort-specific rates of lung cancer from the UPDB were used to estimate the expected number of affected relatives. The cohort-specific lung cancer rate was estimated using all individuals with good genealogy content as the denominator and all lung cancer cases with good genealogy content as the numerator, for each cohort. The observed number of cases among a set of relatives was counted by cohort, excluding duplicates. The expected number of lung cancer cases was estimated by multiplying the number of relatives in each cohort times the cohort-specific rate for lung cancer, then summing overall cohorts. RR was estimated for first-degree, second-degree, third-degree and fourth-degree relatives. The 95% CI for the RR is calculated as in Agresti.20 A p value of less than 0.05 was considered significant for all statistical tests.
Results
Using strict criteria to ensure good genealogy content of at least 12 of 14 immediate ancestors (both parents, all 4 grandparents, and at least 6 of their 8 great grandparents) a smaller set of 1.35 million individuals in the UPDB18 made up the cohort for analysis. There were 657 102 women and 692 207 men; 0.007% are non-white; 1.20 million are born in Utah. These 1.35 million individuals were cross-referenced with the Utah DC and the UCR to identify 291 700 DCs and 93 870 cancer records. Table 1 shows the counts by tobacco contribution code for all deceased lung cancer cases in the UPDB (all cases), as well as for all deceased lung cancer cases with good genealogy content (analysed cases).
Classification of smoking status on a linked death certificate for Utah lung cancer cases
Test for excess relatedness using the GIF method
Results for the GIF tests for excess relatedness among lung cancer cases are shown in table 2 which includes the set of individuals considered (Probands), the number of probands (n), the average relatedness of the probands (Case GIF), the average relatedness of the 1000 sets of controls (Mean control GIF), the empirical significance for the overall GIF (GIF p value), the average relatedness of the probands ignoring close relationships (Case dGIF), the average distant relatedness of the 1000 control sets (Mean control dGIF) and the empirical significance for the dGIF test (dGIF p value). Figure 1 shows the contribution to the GIF statistic by pairwise genetical distance for all lung cancer cases compared with 1000 sets of matched controls, allowing consideration of where the case and control relatedness distributions differ. All lung cancer cases show overall excess clustering over expected (Case GIF 5.30 vs Mean control GIF 4.69; p<0.001); however, as seen in figure 1, most of the excess relatedness is observed for genetical distance 1 (parent/offspring) and 2 (primarily siblings), relationships that share common environment and genetics. The dGIF test for all lung cancers was not significant for all lung cancer cases (Case dGIF 4.23 vs Mean control dGIF 4.11; p=0.057).
Genealogical index of familiality (GIF) analysis for excess relatedness for all lung cancer cases and for subsets of cases by smoking status from death certificate
GIF analysis for all lung cancer cases compared with 1000 sets of matched controls. GIF, genealogical index of familiality.
When only considering non-smoking lung cancer cases (table 2; n=784), significant excess relatedness was observed for all relationships (Case GIF 5.70 vs Mean control GIF 4.71; p<0.001), significant excess relatedness was also observed when close relationships were ignored (Case dGIF 4.62 vs Mean control dGIF 4.11; p=0.020). Significant excess clustering was even observed when only relationships more distant than second cousins once removed (greater than genetical distance 7) were considered (data not shown; p=0.023). Excess relatedness was observed for non-smoking lung cancer cases out to genetical distance 8, third cousins and beyond (figure 2).
GIF analysis for 784 non-smoking-related lung cancer deaths compared with 1000 sets of matched deceased Utah controls. GIF, genealogical index of familiality.
When only smoking lung cancer cases were analysed (table 2; n=1747), overall excess relatedness was significant for all relationships (Case GIF 5.67 vs Mean control GIF 4.74; p<0.001), but when close relationships were ignored, no significant excess relatedness was seen (Case GIF 4.32 vs Mean control GIF 4.18; dGIF p=0.161). Figure 3 shows the GIF results for the smoking-related lung cancer cases compared with 1000 sets of matched deceased Utah controls.
GIF analysis for 1747 smoking-related lung cancer deaths compared with 1000 sets of matched deceased Utah controls. GIF, genealogical index of familiality.
Relative risks in relatives
Table 3 shows estimated RRs for lung cancer for first-degree to fourth-degree relatives of lung cancer cases, including the type of relative (Relative), the number of relatives (n), the observed (obs) and expected (exp) number of relatives with lung cancer, the RR (95% CI), and the significance of the test for increased risk (p value). First-degree, second-degree, third-degree and fourth degree relatives of lung cancer cases are at a significantly increased risk of lung cancer (table 3). These estimates of RR in relatives of lung cancer cases showed similar results to the GIF analysis and support a contribution from genetical and environmental factors. However, degree of relationship and genetical distance do not exactly represent the same measurement. In figure 1, genetical distance 1 and 2 are primarily first-degree relationships, genetical distance 3 is primarily second-degree relationships, genetical distance 4 is primarily third-degree relationships, and genetical distance 5 is primarily fourth-degree relationships.
Estimates of relative risk (RR) for all lung cancers in relatives of all lung cancer cases
The effect of shared environment in lung cancer risk can be examined by estimating the RR for lung cancer in the spouses of lung cancer cases (table 4). RR for lung cancer in the spouses of all lung cancer cases was significantly elevated (RR=2.38; 95% CI 1.81 to 3.06). The RR for lung cancer in the spouses of smoking-related lung cancer cases was also significantly elevated and was even higher (RR=2.75; 95% CI 1.66 to 4.30). Spouses of non-smoking lung cancer cases were the only spouses who did not have significantly elevated risk for lung cancer (RR=1.24; 95% CI 0.50 to 2.55).
Estimates of relative risk (RR) of lung cancer in the last spouse of cases with lung cancer
Table 5 shows the RR for non-smoking-related lung cancer in relatives of non-smoking-related lung cancer probands. For all degrees of relationship analysed, the RR of dying from non-smoking-related lung cancer was elevated, and RRs were significantly elevated for first-degree, second-degree and fourth degree relatives.
Relative risk estimates for non-smoking-related lung cancer among the relatives of 784 non-smoking-related lung cancer probands
Table 6 shows the RR for smoking-related lung cancer among the relatives of smoking-related lung cancer probands. For all degrees of relationship analysed the RR of dying from smoking-related lung cancer was significantly elevated in the relatives of the smoking-related lung cancer cases. All smoking-related lung cancer RRs (table 6) were higher than the RR estimated for all lung cancer cases considered together (table 3).
Relative risk estimates for smoking-related-lung cancer among relatives of 1747 smoking-related probands
Discussion
A number of publications since 196321 support a genetical contribution to lung cancer risk, similar to what has been shown for breast, colon and prostate cancers.22–24 The confounding of shared genetics with shared environmental exposure (eg, smoking, occupation, childhood environment) makes separation of the genetical contribution to lung cancer challenging. Recognised comorbid conditions such as COPD are independent risk factors regardless of smoking status.25 Additionally, unrecognised factors such as environmental tobacco smoke, radon, occupational exposures and diet may contribute to the development of lung cancer in smokers and non-smokers.5 ,17 ,26 ,27 This unique study of the familial clustering of lung cancer using a population-based resource linking genealogy, a tumour registry and DC data begins to clarify the role of genetics in non-smoking-related lung cancer.
The GIF results clearly show significant excess relatedness for close and distant relationships for non-smoking-related lung cancer cases. Additionally, significantly increased RR for non-smoking-related lung cancer cases among close and distant relatives is shown. These results are the first strong support for a genetical contribution to the subset of non-smoking-related lung cancer.
GIF analysis of the subset of smoking-related lung cancers also shows evidence for significant excess relatedness, but the dGIF results show excess familial relatedness observed was primarily due to close relationships; it was not observed when ignoring close relationships. The RR test differs from the GIF as it estimates risk for specific sets of relatives. The RR results for the smoking-related set of cases show significantly increased risks out to fourth-degree relatives, supporting a strong possibility that a genetical predisposition in some cases is enhanced by the additional risk factor of smoking.
Shared environmental exposure, specifically environmental tobacco smoke, has previously been associated with lung cancer.28 In a meta-analysis an OR of 1.26 (95% CI 1.07 to 1.47) was seen in non-smoking spouses of smokers and there was a linear relationship between lung cancer risk and the quantity and duration of exposure of smoking by the partner.26 We demonstrate the RR for lung cancer in the spouses of smoking-related lung cancer cases (RR=2.75; 95% CI 1.66 to 4.30) is similar to the RR of smoking-related lung cancer in first-degree relatives of smoking-related lung cancer cases (RR=2.58; 95% CI 2.13 to 3.10), suggesting genetical risk factor(s) may be equivalent in strength to the smoking risk factor.
The possibility of a synergistic effect between smoking and genetics29 is certainly likely. While the aim of this analysis was to explore evidence for a genetical contribution to non-smoking-related lung cancer, perhaps the most intriguing results are the RR estimates for smoking-related lung cancers. RR analysis differs from the GIF analysis as it only considers one relationship at a time, while the GIF analysis considers all pairs of cases. RR estimates for smoking-related lung cancer in the deceased relatives of smoking-related lung cancer cases were the highest RR observed (table 5), and were significantly elevated out to fourth-degree relatives. We hypothesise this may represent evidence for a gene-environment interaction in individuals who carry a predisposition variant, and who have further increased risk based on smoking behaviour. Genome-wide association studies support evidence for such a gene-environment interaction.30 However, the effect of the genetical variant or familial mutation may be muted if the environmental exposures were absent in past generations.31 Until the responsible predisposition genes are identified, and we can observe the outcomes in smoking and non-smoking predisposition carriers, we may not fully understand these combined effects.
In the most similar published study, Jonsson et al6 evaluated the familial risk of lung cancer in the Icelandic population. In Iceland, as in Utah, there is a genealogical database and records are available for cause of death since 1955. A significantly increased risk of lung cancer was observed in first-degree, second-degree and third-degree relatives, with RRs of 2.69 (95% CI 2.20 to 3.23), 1.34 (95% CI 1.15 to 1.49) and 1.28 (95% CI 1.10 to 1.43), respectively. These RRs are similar to those observed for all lung cancer cases in Utah for first-degree, second-degree and third-degree relatives, respectively: 2.18 (95% CI 2.03 to 2.34), 1.33 (95% CI 1.23 to 1.43) and 1.13 (95% CI 1.08 to 1.18). One limitation of the Icelandic study was that smoking information was only available for about 20% of their cohort, and direct adjustments for smoking was not possible. This is important when comparing it with this Utah study, as the prevalence of smoking in Iceland was over 20% in 2004,32 which was considerably higher than Utah's estimate of 10%.33
There are limitations to analysis of a resource like the UPDB. These limitations include censorship, quality of DC data and lack of data for other known environmental risk factors such as secondhand smoke or occupation. Data for individuals who were not part of the original pioneer families or who do not have Utah vital statistics data would not be included in the genealogy and could therefore not be linked to cancer and DC data. Cancer data for individuals diagnosed out of state, or before 1966, would not be included. Tobacco use data was not available for lung cancer cases still alive, for those who died outside Utah, or for those whose attending physician at death did not complete the tobacco-related questions. While the DC has separate codes for the contribution of tobacco to the final cause of death, it relies on the physician to provide these data. These limitations apply to the entire UPDB, including cases and controls; they are assumed to be uniform and not associated with bias that would affect the hypothesis tests performed, but may reduce the power of these analyses.
While the population of Utah is very representative of the Mormon founding population from the mid-1800s, it does not represent an isolated population. The pioneer founders were a largely unrelated group of about 20 000 northern Europeans from England, Scotland, Wales, Denmark and Sweden.34 Migration over ensuing generations contributed an additional 30 000–50 000 people to Utah. Recently, there has been a substantial influx of Hispanic, Polynesian and international religious migrants, but genealogies for these may not be complete in the UPDB. Utah has low to normal levels of inbreeding, compared with the rest of USA.35 The population of Utah is recognised to be representative of USA and northern Europe; however, extrapolations to any other populations should not be made without additional analyses.
In this unique analysis, existing population-based data, with relationship data known for up to 12 generations, was linked to a state-wide cancer registry and DC data to explore the relationships between environment and genetics in the development of lung cancer. We show strong supporting evidence for a genetical contribution to non-smoking-related lung cancer while confirming the well-recognised evidence for familial clustering of smoking-related lung cancers. The failure of prior efforts to identify lung cancer predisposition genes of interest may in part be due to a focus on pedigrees with closely related, smoking-related lung cancers in whom a genetical predisposition gene is not always present. We propose screening and identification efforts for lung cancer predisposition genes should focus on the subset of high-risk non-smoking-related lung cancer pedigrees to lessen the noise from tobacco's contribution to lung cancer.
References
Footnotes
Contributors All authors contributed to the design. SRC and LAC-A were responsible for acquiring the data, interpretation of the data, drafting and editing the manuscript, while WA and MH additionally contributed in the editing of the manuscript. All authors have approved the final revised draft of the manuscript.
Funding This work was supported by the Utah Cancer Registry, which is funded by Contract No. HHSN261201000026C from the National Cancer Institute’s SEER Program with additional support from the Utah State Department of Health and the University of Utah. Partial support for all data sets within the Utah Population Database (UPDB) was provided by Huntsman Cancer Institute, University of Utah and the Huntsman Cancer Institute’s Cancer Center Support grant, P30 CA42014 from National Cancer Institute. LAC-A acknowledges support from the University of Utah Center on Aging and the Huntsman Cancer Foundation.
Competing interests None declared.
Ethics approval Institutional Review Board.
Provenance and peer review Not commissioned; externally peer reviewed.
Linked Articles
- Airwaves